Extensive microperfusion system (EMPS) for bacterial population experiments
Microfluidic devices and microbial experiments
Microbial experiments using microfluidic devices have been widely conducted in recent years [A]. Microfluidic devices typically consist of microchannels created by microfabrication technologies used in integrated circuit fabrication processes. By constantly feeding fresh media into the flow channel, one can culture various microorganisms under designed and controlled environments, and carry out observations at both single-cell and collective levels. Therefore, microfluidic devices are a very useful tool for understanding biological phenomena by physical approaches, that is by designing and studying simplest possible problems that capture the essence, through quantitative characterization of controlled experiments.
In our Lab, from the viewpoint of statistical physics and related areas, we are trying to understand cooperative behavior of dense bacterial populations. For example, dense clumps of bacteria seen in kitchen sinks, gums, and medical equipments are called biofilms and found everywhere in our daily lives. Within a biofilm, cells communicate with each other by mechanical interactions as well as via signaling molecules, and the whole population occasionally exhibits cooperative behavior [B]. We observe such dense bacterial populations in controlled environments and aim to understand what kind of collective properties arises in bacteria, given that their single-cell behavior is relatively well understood.
A new microfluidic device: extensive microperfusion system (EMPS)
Although microfluidic devices have been used for various studies on microorganisms, it has been difficult to measure dense bacterial populations for a long time in a spatially uniform environment. Commonly used microfluidic devices consist of channels made of a silicone elastomer called PDMS (polydimethylsiloxane), through which culture medium is supplied. In the case of dense bacterial populations, nutrients are exhausted by bacteria near the inlet and do not reach farther regions. Therefore, the culture condition becomes spatially heterogeneous.
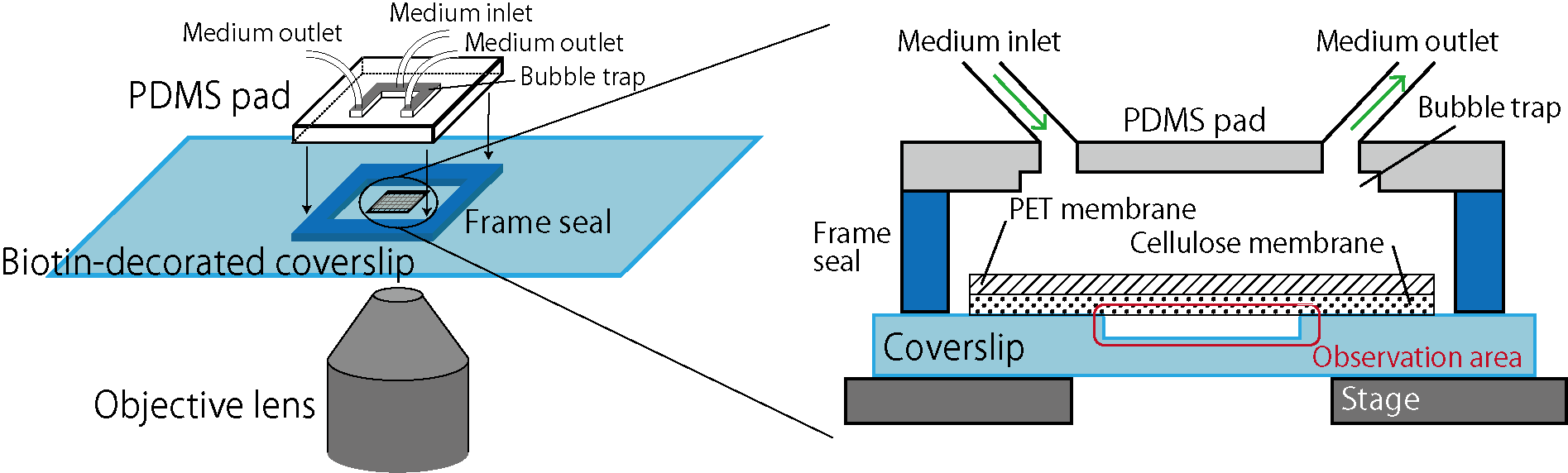
Here we focus on membrane-based microfluidic devices and developed a new system for controlled experiments of dense bacterial populations (Fig.1). Wells are designed and fabricated on a coverslip by microfabrication technology. Bacterial cells are confined in those wells, covered by a porous membrane from the top using biotin-streptavidin binding (biocompatible adhesion). This realizes spatially uniform control of the culture environment by medium exchanges through the porous membrane while bacteria cells are kept trapped in the wells. The previous system [C] used cellulose as a porous membrane, but because this membrane is soft and bends, it was difficult to realize an extended observation area. Here, we reduced the membrane bending by reinforcing the cellulose membrane by a rigid PET (polyethylene terephthalate) porous membrane, and thereby we realized controlled experiments of bacterial populations in extended areas [1]. We name this device the extensive microperfusion system (EMPS) and expect that it will be a useful platform for various controlled and quantitative experiments involving cell populations.
We are now working on various research subjects on bacterial populations, using the EMPS. Those include statistical laws of cell size distributions in E. coli populations, crowding effect of bacterial populations, and competition dynamics of multiple populations, and we also plan to work on biofilms.
References
(Takeuchi Lab)
[1] T. Shimaya, R. Okura, Y. Wakamoto and K. A. Takeuchi, Communications Physics 4, 238 (2021) [web].
(other groups)
[A] G. Velve-Casquillas, M. L. Berre, M. Piel, and P. T. Tran, Nano Today 5, 28 (2010) [web].
[B] L. Hall-Stoodley, J. W. Costerton and P. Stoodley, Nat. Rev. Microbiol. 2, 95 (2004) [web]; H. Boudarel, J. Mathias, B. Blaysat and M. Grédiac, npj Biofilms Microbi. 4, 17 (2018) [web]; C. Fuqua, A. Filloux, J. M. Ghigo and K. L. Visick, J. Bacteriol. 201, e00118 (2019) [web].
[C] I. Inoue, Y. Wakamoto, H. Moriguchi, K. Okano and K. Yasuda, Lab Chip 1, 50 (2001) [web].
Main contributors
T. Shimaya, K. A. Takeuchi
